Amino
Advertisement
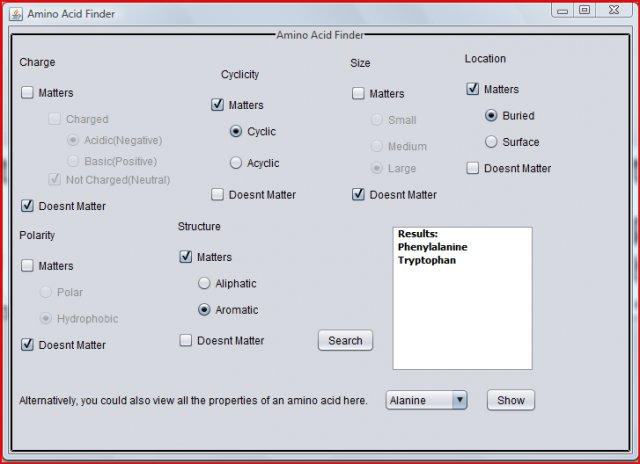
Amino Acid Finder v.1.0
Ever got confused while remembering all the various physical properties of all the standard 20 amino acids?
Advertisement
AmiPig v.1.0
The AmiPig table provides amino acid ileal digestibility values for 62 common feedstuffs used in pig diets.
Genome Explorer v.1.0
Search sequence data with this tool. Genome Explorer offer users a single interface for some useful bioinformatics tools. It provides additional functionality for searching sequence data (for particular groups of nucleotides or amino acids) and
PhyDE v.0.997
View and analyze DNA sequence alignments with this tool. PhyDE (Phylogenetic Data Editor) is a system-independent editor for DNA and amino acid sequence alignments,
STRAP (Structure based Sequences Alignment Program) v.1.0
Protein alignment made easy. STRAP (Structure based Sequences Alignment Program) aligns proteins by sequence and 3D-structure. It supports the simultaneous analysis of hundreds of proteins and integrates amino acid sequence, secondary structure,
DAMBE (Data Analysis and Molecular Biology and Evolution) v.5.2.30
DAMBE (Data Analysis and Molecular Biology and Evolution) help you with data analysis and molecular biology and evolution.Here is a short summary of DAMBE functions 1. Sequence alignment * General sequence alignment with nucleotide and amino acid
Genetic Code v.1.0
Study the genetic code with this tool. Genetic Code can display the universal genetic code, the one- and three-letter amino acid codes, the biochemical structures of the amino acids,
Rate4Site v.2.01
Rate4Site is a software for detecting conserved amino-acid sites by computing the relative evolutionary rate for each site in the multiple sequence alignment (MSA).The rate of evolution is not constant among amino acid sites:
Seq-Gen v.1.3.2
Seq-Gen is a software that will simulate the evolution of nucleotide or amino acid sequences along a phylogeny, using common models of the substitution process.
ProSAT (PROtein reSidue Annotation Toolkit) v.1.0
PROtein reSidue Annotation Toolkit. ProSAT toolkit is a set of programs that allow building SVM based models for annotating amino acid residues in protein sequences using user supplied features (like PSI-BLAST profiles, or PSIPred profiles).
IQPNNI - Important Quartet Puzzling and NNI Operation v.3.3.2
Important Quartet Puzzling and NNI Operation. An efficient tree reconstruction method (IQPNNI) is introduced to reconstruct a phylogenetic tree based on DNA or amino acid sequence data.